flowchart LR
A["app.py"] --- A1("layout页面组件")
A["app.py"] --- A2("callback函数")
A2 --- A21("render_js():加载粒子效果")
A2 --- A22("route():根据url加载不同内容")
%% A21 --> A211("Input('url', 'pathname')")
%% A21 --> A212("Output('execute-js', 'jsString')")
A1 --- A11("dcc.Location(id='url'):监听浏览器的url")
A1 --- A12("fuc.FefferyExecuteJs(id='execute-js'):加载粒子效果的js")
A1 --- A13("html.Div(id='particles-mount'):加载粒子效果的html")
A1 --- A14("html.Div(id='app-mount'):页面内容的容器")
A1 --- A15("html.Div(id='router-redirect-container'):路由重定向的容器,加载dcc.Location,以监听url变化")
A1 --- A16("html.Div(id='index-user-manage-add-user-message-container'):消息容器")
A11 -- 回调 --> A12
A11 -- 回调 --> A14
A11 -- 回调 --> A15
周打卡
2023年10月
第1周
- 【NMPA高研院】您报名的药品GCP课程已开通,请登录网址: https://www.nmpaied.com 使用报名手机号和验证码登录学习。请务必于40天内完成全部课程的学习和考试。
- 15521113267/Gcp@31415926;920500/Asd8569051.@用户名/密码错误
- 考试成绩:88分。82分通过。
- 专业: 内科学(肾脏病学)
- 住培师资:【金山文档】 国家级住院医师规范化培训证书-胡林辉 https://kdocs.cn/l/ctaZwwVl5iyk
- 暨大官网https://www.jnu.edu.cn/main.htm ⇒ 学籍管理 ⇒ 毕结业应用 ⇒ 非学历结业申请 ⇒ 修改姓名拼音、籍贯、政治面貌等需要更新的内容。
- 信息:内招生。
论文:Her2 2023-12-04 16:37:28 CST
-
-
-
Section Content Tables/Figures Included Clinical Characteristics of Patients with HER2 Mutations - Clinical characteristics of patients with HER2 mutations Figure 1, Table 1 Genetic Characteristics of HER2 Mutations - Spatial distribution of ERBB2 mutations in NSCLC
- Co-mutation patterns of patients with concomitant EGFR mutations and HER2 amplification lung cancerSupplementary Table 2, Figure 2, Figure 3 Heterogeneity of HER2 mutations - Comparison of mutation differences in Exon20 and Non-exon20 regions of HER2
- Variation analysis of gene mutations between Baseline and Non-baseline samples- Figure 6, Supplementary Table 2
-Figure 5, Figure 4, Summplementary Table 1In this adjustment, the figures are reordered to create a more coherent and logical progression of the research findings, ensuring that the presentation of results aligns with the narrative flow.
-
GO enrichment analysis的描述参考 https://www.frontiersin.org/articles/10.3389/fendo.2023.1213465/full
-
-
概括研究结果及临床意义,本研究的创新点。
与前人研究做比较,有哪些新的发现或补充。
本研究的优点与缺点。
-
- Step 1: Adding references to your project
- Step 2: Synchronize your R Markdown document with CiteDrive
- Step 3: Copy citation keys
-
Declarations @交给老婆
2023-11-14 01:03:25 CST -
-
-
- 生成表格的工具:https://tablesgenerator.com/
- 手动锚点的方法:
<a href="#tbl-table1">Table 1</a>
Clinicopathologic Characteristics of the EGFR Gene Mutation in Non–small Cell Lung Cancer A Prospective Cohort Study to Define the Clinical Features and Outcome of Lung Cancers Harboring HER2 Aberration in Japan (HER2-CS STUDY)
Comparisons of normally distributed data were performed using Student’s t tests, and non-normally distributed data using Wilcoxon rank sum tests. Comparisons of categorical variables were performed using Fisher’s exact tests, P value <0.05 was considered significant. Where applicable, P values were adjusted for multiple group comparisons by Bonferroni’s post-hoc test or corrected for multiple hypotheses testing using the false discovery rate (FDR) adjustment method as appropriate. Adjusted P value <0.1 was considered significant. For survival analyses, Kaplan-Meier curves were compared using the log-rank test, and hazard ratios (HRs) were calculated by Cox proportional hazards model. All statistical analyses were done in R (v.3.5.2).
- 论文排布实例
- 相似文献
Real World Characteristics and Clinical Outcomes of HER2-Mutant Non–Small Cell Lung Cancer Patients Detected by Next-Generation Sequencing PDF
Clinicopathologic Characteristics of the EGFR Gene Mutation in Non–small Cell Lung Cancer
- 文献支持
Clinical Characteristics and Prognosis of HER2 Gene Phenotype in Patients with Non-Small Cell Lung Cancer
Treatment outcome and clinical characteristics of HER2 mutated advanced non-small cell lung cancer patients in China
Clinical Characteristics and Outcomes of Non-small Cell Lung Cancer Patients with HER2 Alterations in Korea
A Prospective Cohort Study to Define the Clinical Features and Outcome of Lung Cancers Harboring HER2 Aberration in Japan (HER2-CS STUDY) Clinicopathologic Characteristics of the EGFR Gene Mutation in Non–small Cell Lung Cancer
Lung Cancer That Harbors an HER2 Mutation: Epidemiologic Characteristics and Therapeutic Perspectives
- 表
【金山文档】 Tables-her2 https://kdocs.cn/l/crTgL8NH7OUk
- 图
【金山文档】 Figures-her2 https://kdocs.cn/l/cdGuiNdMvOYE
我是在这个网站做的图:http://www.bioinformatics.com.cn/plot_basic_go_pathway_circlize_plot_140
- 辅助材料
方法学:【金山文档】 Geneseeq methods for NGS panel https://kdocs.cn/l/caQs4Zou5PF5
不知来源的文本
Targeted RNA and DNA sequencing were done using TruSight Tumor 170 (Illumina, San Diego, CA). The TruSight Tumor 170 panel was designed to detect 170 cancer-related genes, including 151 genes with potential single nucleotide variants and indels, 59 with potential amplifications, and 55 with fusion and splice variants (S2 Table). Briefly, 40 ng of formalin-fixed paraffin-embedded (FFPE) tissue-derived DNA and RNA samples were extracted using a QIAGEN AllPrep DNA/RNA FFPE Kit (Qiagen, Hilden, Germany). After hybridization capture-based target enrichment, pairedend sequencing (2×150 bp) was performed on a NextSeq sequencer (Illumina) according to the manufacturer’s instructions. Variants with a total depth of > 100× and a variant allele frequency of > 3% were included for analysis. Variant interpretation was based on the recommendations of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists [10] (Supplementary Methods). Actionable genetic alterations were stratified into one of four levels based on the OncoKB website (http://www.OncoKB.org). Tier 1 variants, which include level 1 and level 2 genetic alterations, are Food and Drug Administration (FDA)–approved biomarkers and the standard of care, whereas tier two variants included alterations with compelling clinical or preclinical evidence to drug response.
- 信息更新
数据没有结局,故在标题和摘要均删除结局
临床信息.xlsx
2022-11-11 NSCLC HER2突变信息汇总-LCQ调整结果.pptx
ex20GO_Three_Ontologies-中文.pdf
non20GO_Three_Ontologies-中文.pdf
ex20GO_Three_Ontologies-中文_00.png
non20GO_Three_Ontologies-中文_00.png
ERBB2_lollipop (1).png
饼图.pdf
oncoprint-all.genelist-combined.pdf
ex20 GO Top10.pdf
non20 GO Top10.pdf
oncoprint-baseline.top30.pdf
oncoprint-all.top30-all.pdf
金山文档
【金山文档】 2022-11-11 NSCLC HER2突变信息汇总-LCQ调整结果 https://kdocs.cn/l/ce888O5Wne7s
【金山文档】 临床信息 https://kdocs.cn/l/cgZGd3dQCBx7
【金山文档】 ERBB2_lollipop2 更新版 https://kdocs.cn/l/cljxZzLycmJg图的解读
BMC参考图
| 名称 | 图片 | 图注 |
|---|---|---|
| erbb2-mutation |  |
Figure 1. Rate of ERBB2 and ERBB3 mutations and their spatial distribution on HER2/3 in ILC and IDC. ERBB2mut were found to be a enriched in ILC (yellow bar) vs. IDC (blue bar) and b clustered in the tyrosine kinase domain of HER2; c ERBB3mut occurred at lower frequency, with the high-frequency outlier in IDC coding for known oncogenic HER3 kinase domain alteration E928G (N = 6). Y-axes show the number of cases harboring at least one ERBB2/3 mutation at a specific amino acid (aa) of HER2/3, shown along the x-axes. Yellow-filled circles indicate oncERBB2mut. Extracellular domains of HER2/3: Receptor L, Furin-like and Growth Factor Receptor IV; intracellular: tyrosine protein kinase. p < 0.001; n/s = not significant ERBB2和ERBB3突变在乳腺导管癌(IDC)和乳腺小叶癌(ILC)中的发生率及其在HER2/3上的空间分布。ERBB2突变在ILC(黄色柱状图)相比于IDC(蓝色柱状图)的富集情况,并且聚集在HER2的酪氨酸激酶结构域中;ERBB3突变的发生频率较低,在IDC中存在一个高频突变(编码已知致癌性HER3酪氨酸激酶结构域改变E928G,有6例)。Y轴表示在HER2/3的特定氨基酸位置上携带至少一个ERBB2/3突变的案例数,X轴显示相应的氨基酸位置。黄色填充圆表示具有致癌性ERBB2突变。HER2/3的细胞外结构域包括受体L、Furin-like和生长因子受体IV;细胞内结构域包括酪氨酸蛋白激酶。p<0.001;n/s表示不显著。 |
| ERBB2_lollipop | 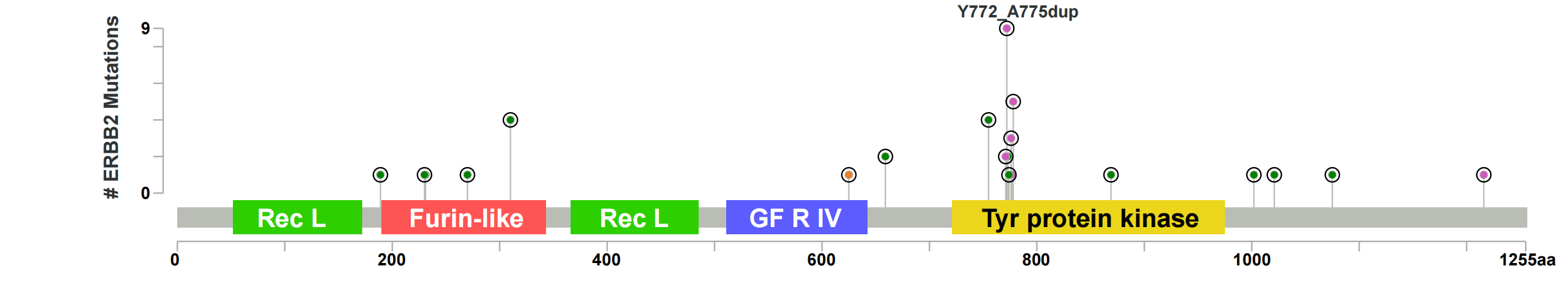 |
ERBB2 mutations and their spatial distribution on HER2 in NSCLC. ERBB2mut were found to be clustered in the tyrosine kinase domain of HER2; Y-axes show the number of cases harboring at least one ERBB2 mutation at a specific amino acid (aa) of HER2, shown along the x-axes. Yellow-filled circles indicate oncERBB2mut. Extracellular domains of HER2: Receptor L, Furin-like and Growth Factor Receptor IV; intracellular: tyrosine protein kinase. 结果解读:共61例样本检出32种ERBB2的变异形式,包括8种20ins突变,其中ERBB2 p.Y772_A775dup 检出最多,分别占基线和非基线的40%和41.7%。 |
| 原文 | 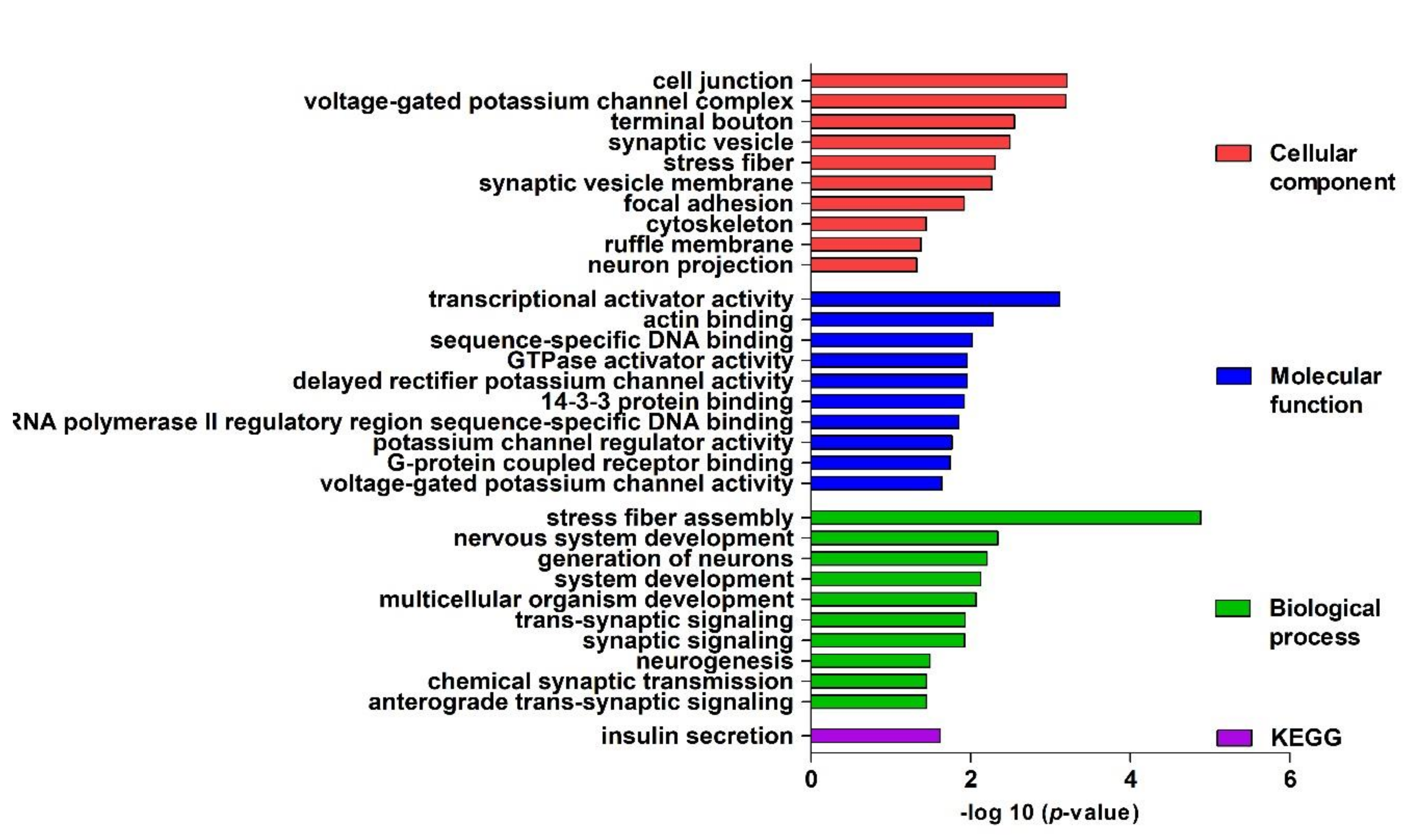 |
Figure 2. Gene ontology (GO) enrichment analysis. Top 10 significantly enriched GO (−log10 (p-value)) terms of the target genes in the cellular components, molecular function, and biological processes. KEGG, Kyoto Encyclopedia of Genes and Genomes. |
| 原文 | 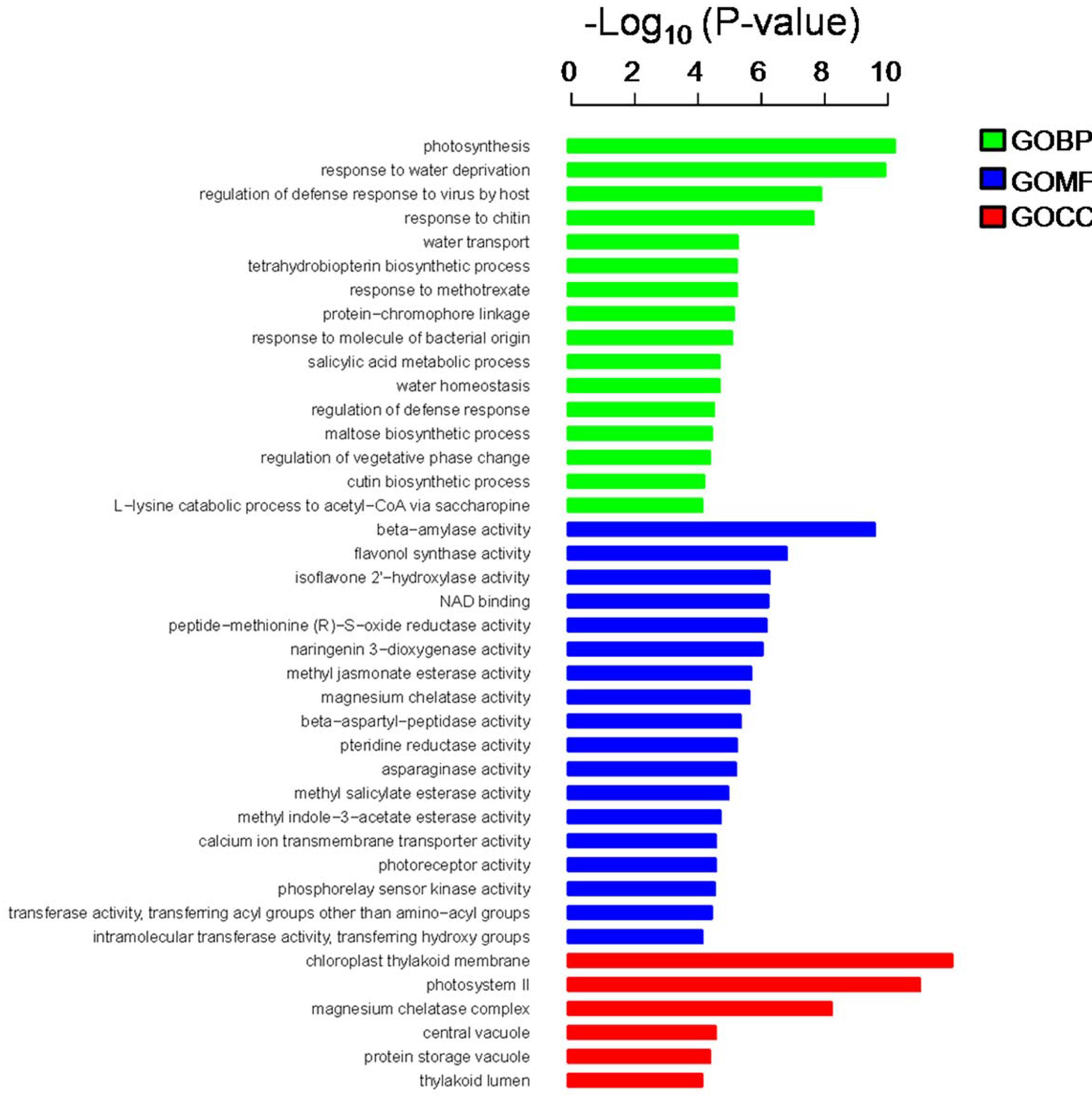 |
FIGURE 3. Gene ontology (GO) annotation and enrichment analysis of 947 DEGs. The horizontal axis shows the negative log10 of the p-value, while the vertical axis represents biological process, molecular function, and cellular component, respectively. |
| ex20GO_Three_Ontologies-中文_00.png | 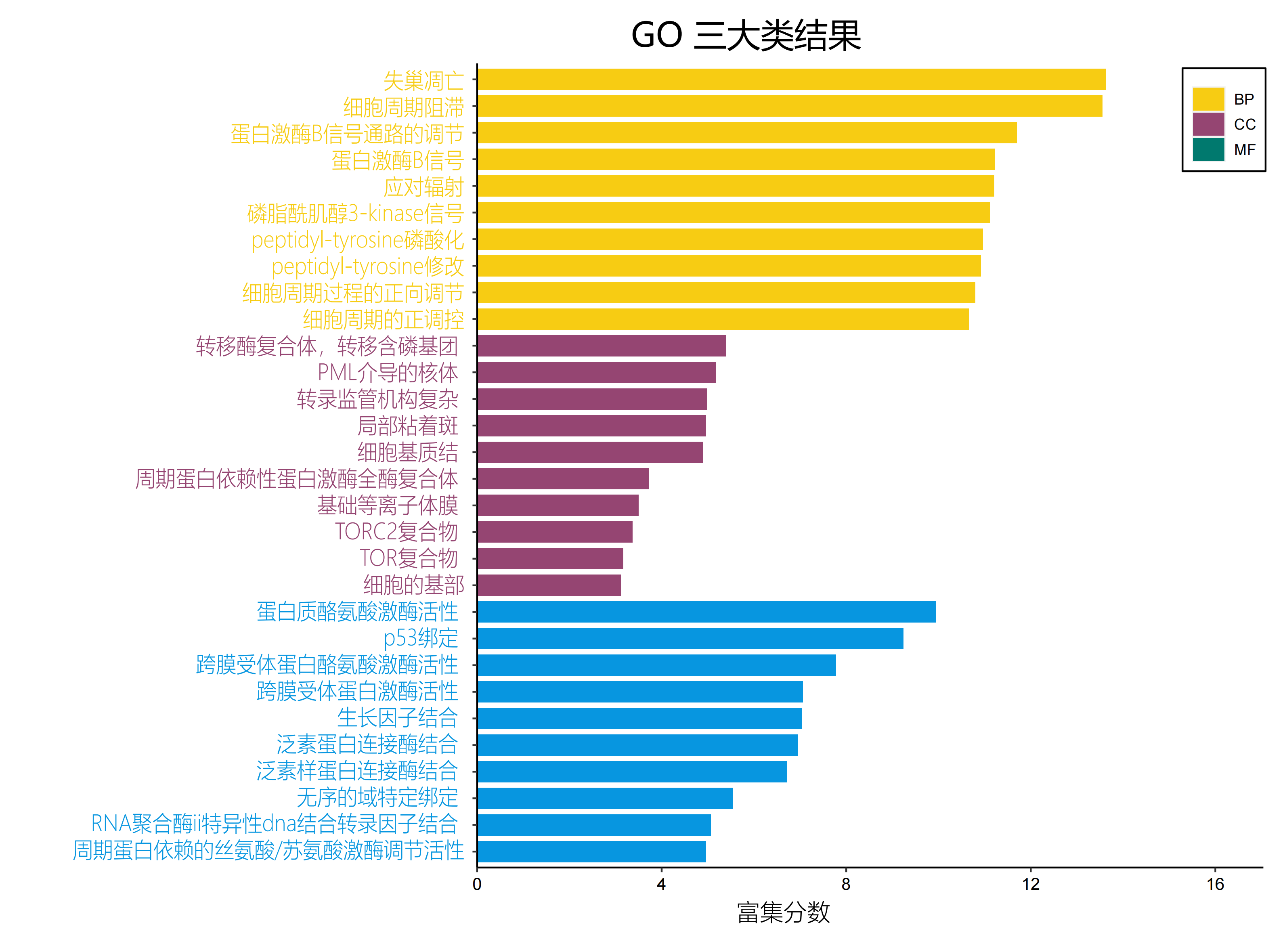 |
有没有英文图? |
| non20GO_Three_Ontologies-中文_00.png | 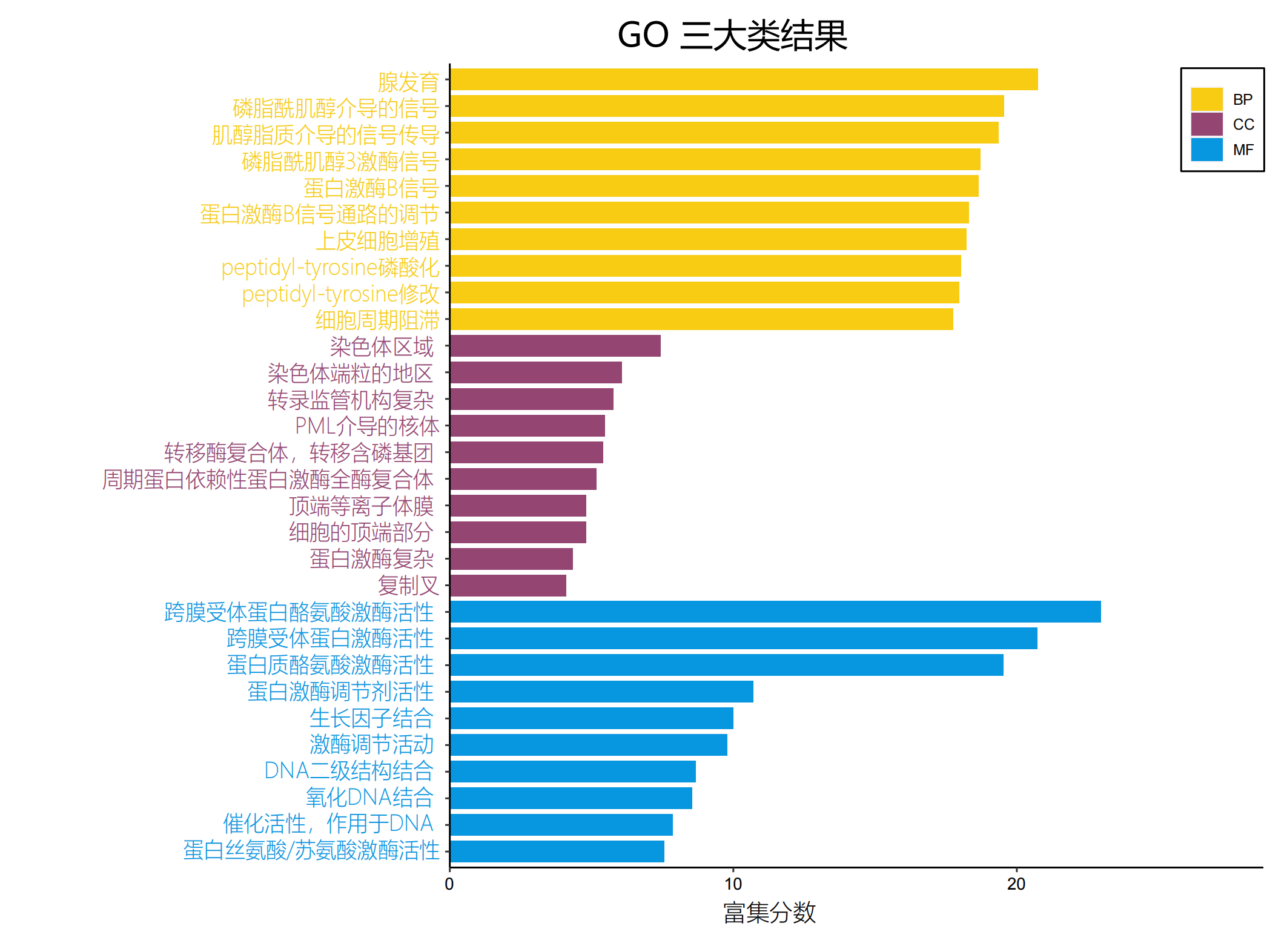 |
有没有英文图? |
| 饼图 | 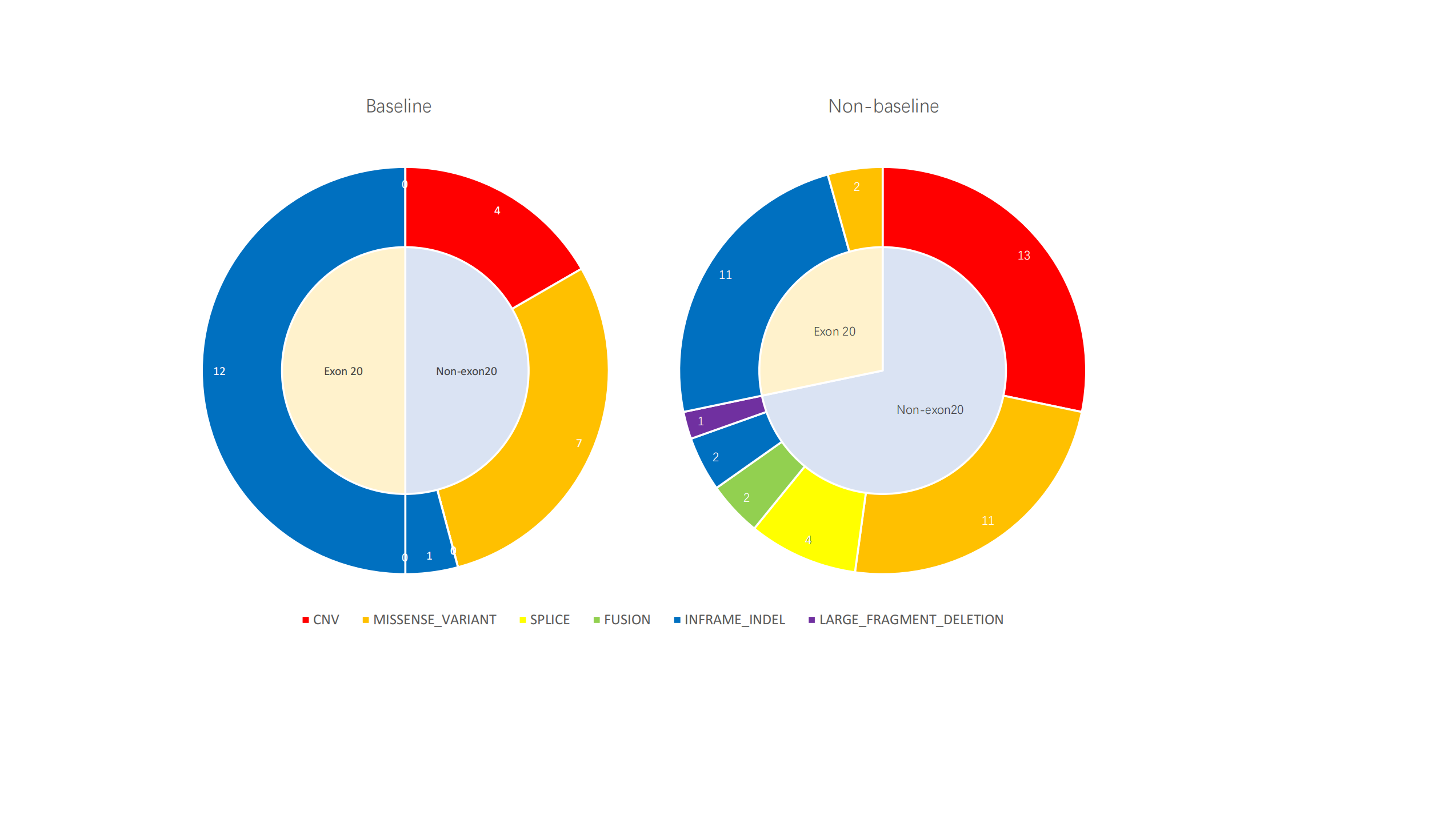 |
基线样本发生在20号外显子上的ERBB2突变100%为Ex20ins 除Ex20ins突变外,拷贝数变异为ERBB2的主要变异形式 非基线样本中有28%为ERBB2拷贝数变异 基因CNV的扩增与缺失,在基线与非基线样本中分布均衡(扩增vs缺失 基线2vs2,非基线 6vs7) NON EX 20主要是CNV和missense variant为主,用药后改变? 可以这么分析,EGFR-TKI的使用可能会导致ERBB2拷贝数变异,而ERBB2扩增也已证明是EGFR-TKI的重要耐药机制之一 |
| oncoprint-all.genelist-combined_00.png | 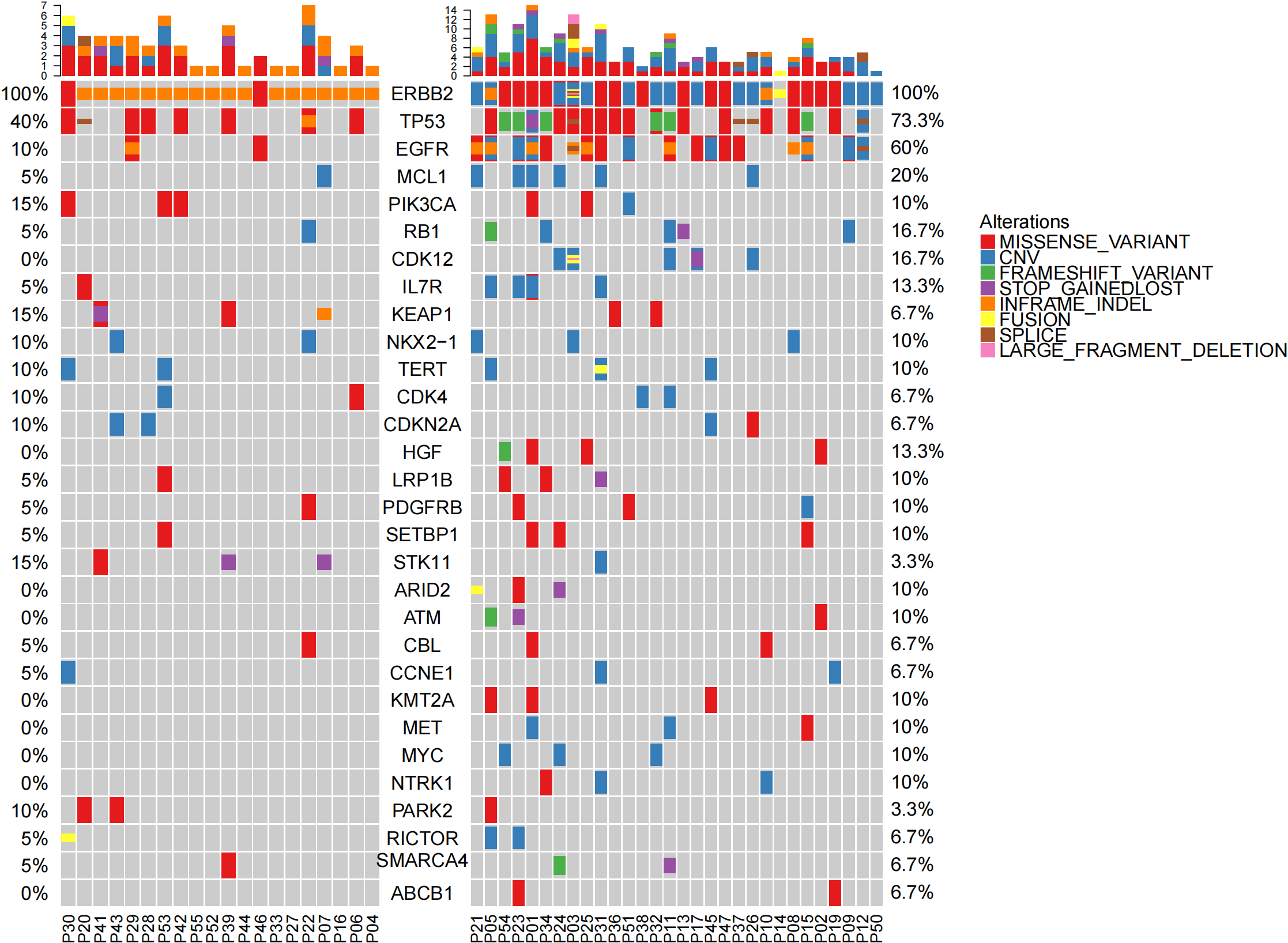 |
EGFR及TP53在non-ex20组患者中显著增加 (Supplementary Table 2) ERBB2 20ins中有2例EGFR阳性,突变形式包括L858R(P46)/19del和L861Q(P29) 既往报道ERBB2 20ins突变与EGFR互斥。 |
| oncoprint-baseline.top30_00.png | 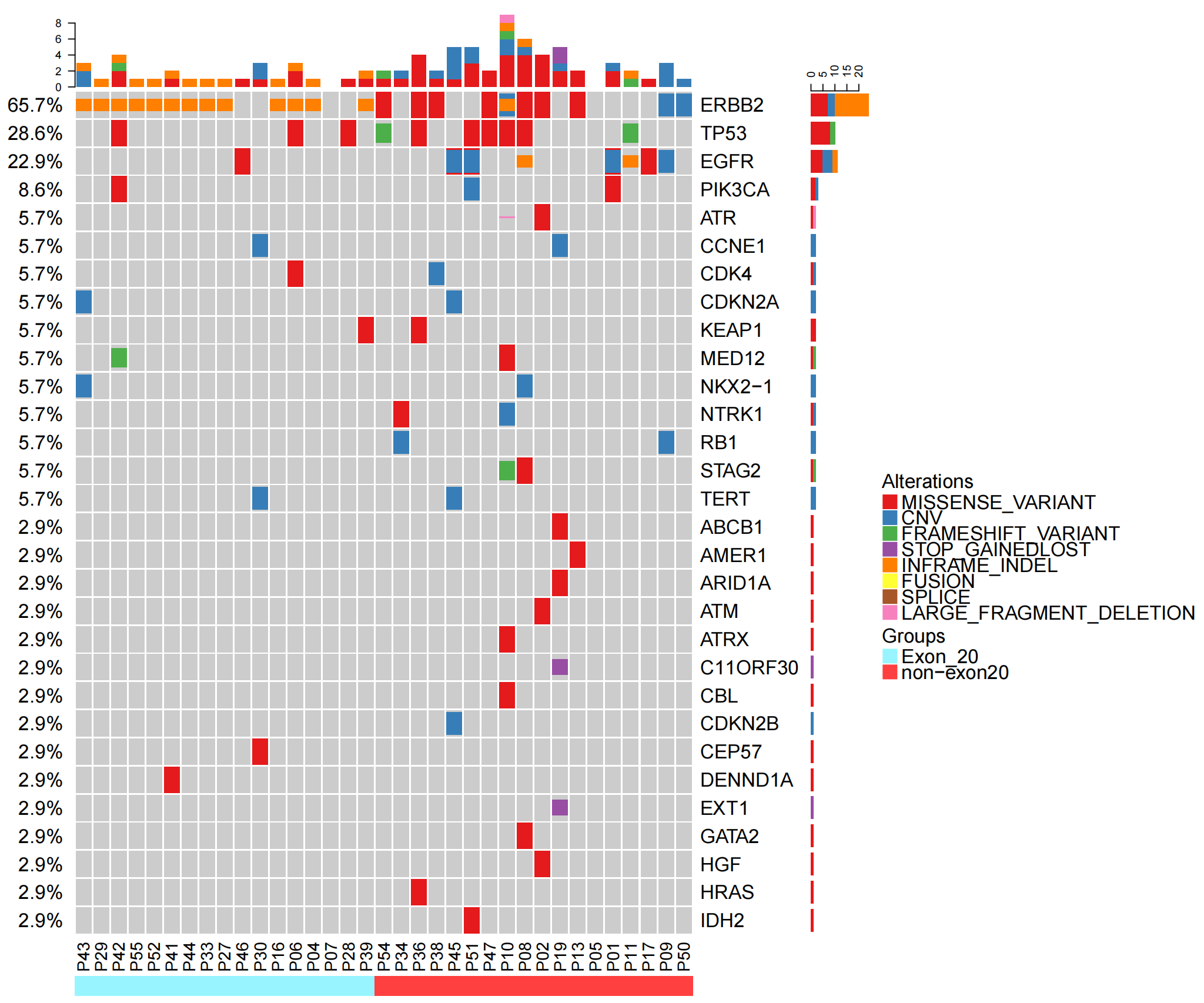 |
35例患者的基线样本中65.7%ERBB2阳性。基线样本的定义?未经任何全身性抗肿瘤治疗的样本。具体为采样日期在确诊日期前后1周,及结合有记录的治疗史。由于临床数据不全,结果可能有偏差。 EX-20只有17个病例,NON EX-20 有18个,总共35个?仅有这些患者有(可判断的)基线样本 Erbb 2继发于EGFR? 可以这么猜想 |
| oncoprint-all.top30-all_00.png | 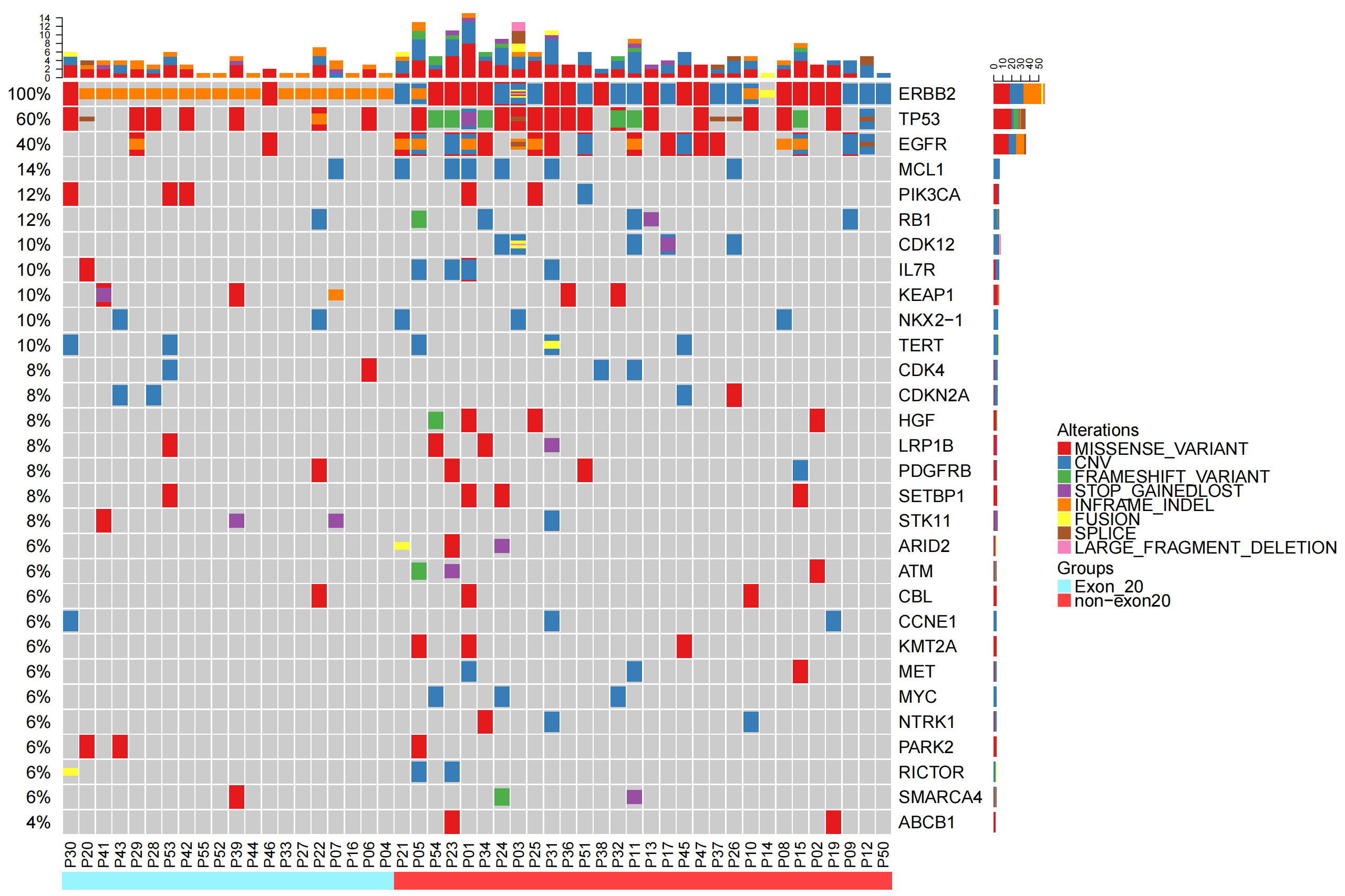 |
Exon 20突变组与non-Exon20突变组的突变图谱有差异 |
| ex20 GO Top10_00.png | 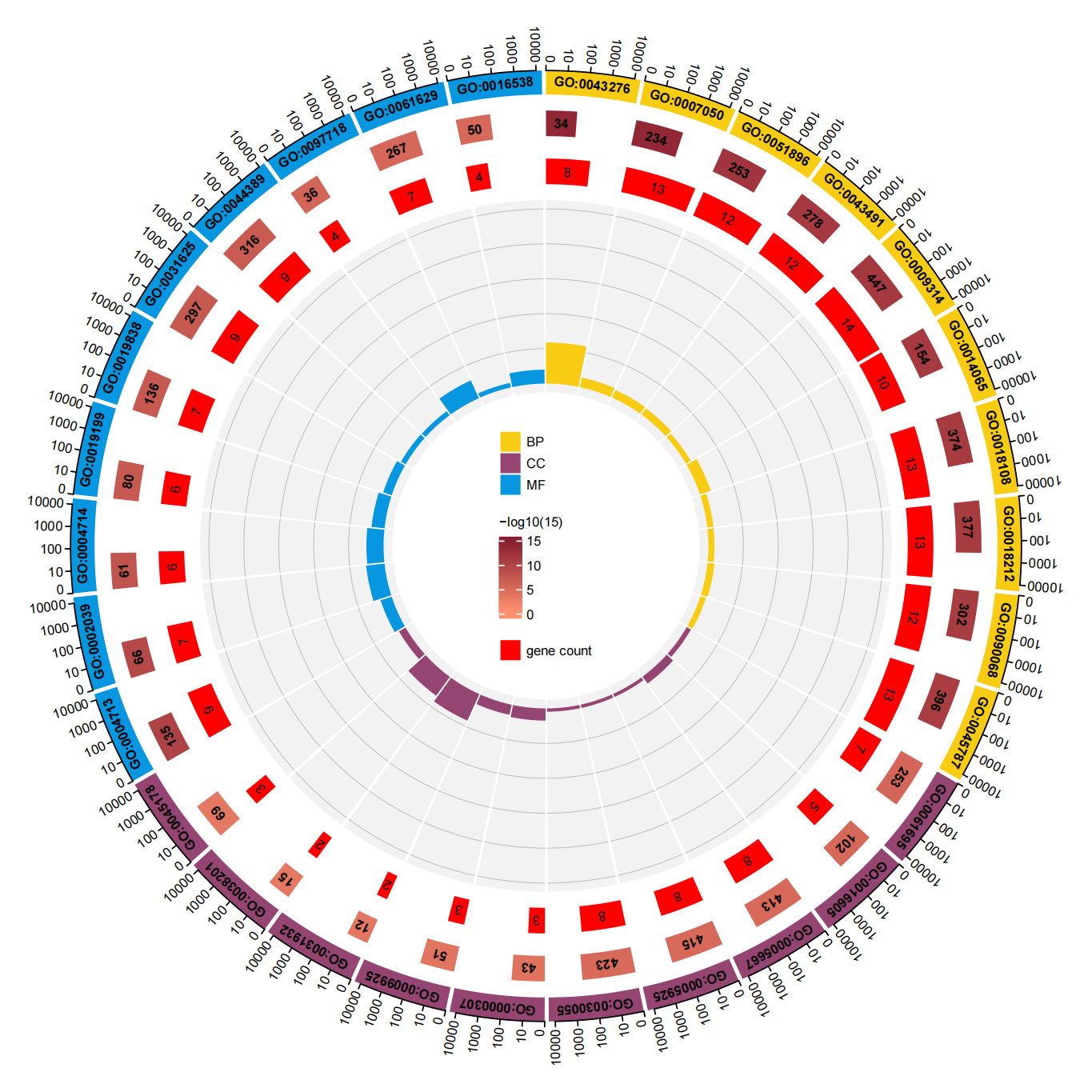 |
使用圆形展示富集结果。从外到内依次为: GO分类以及鉴定到的GO ID 数字:在该GO分类中的基因总数,对应最外的刻度,颜色:深浅代表富集的显著性在该通路下样本富集的基因数 rich factor(第3通道基因数/第2通道基因数) |
| non20 GO Top10_00.png | 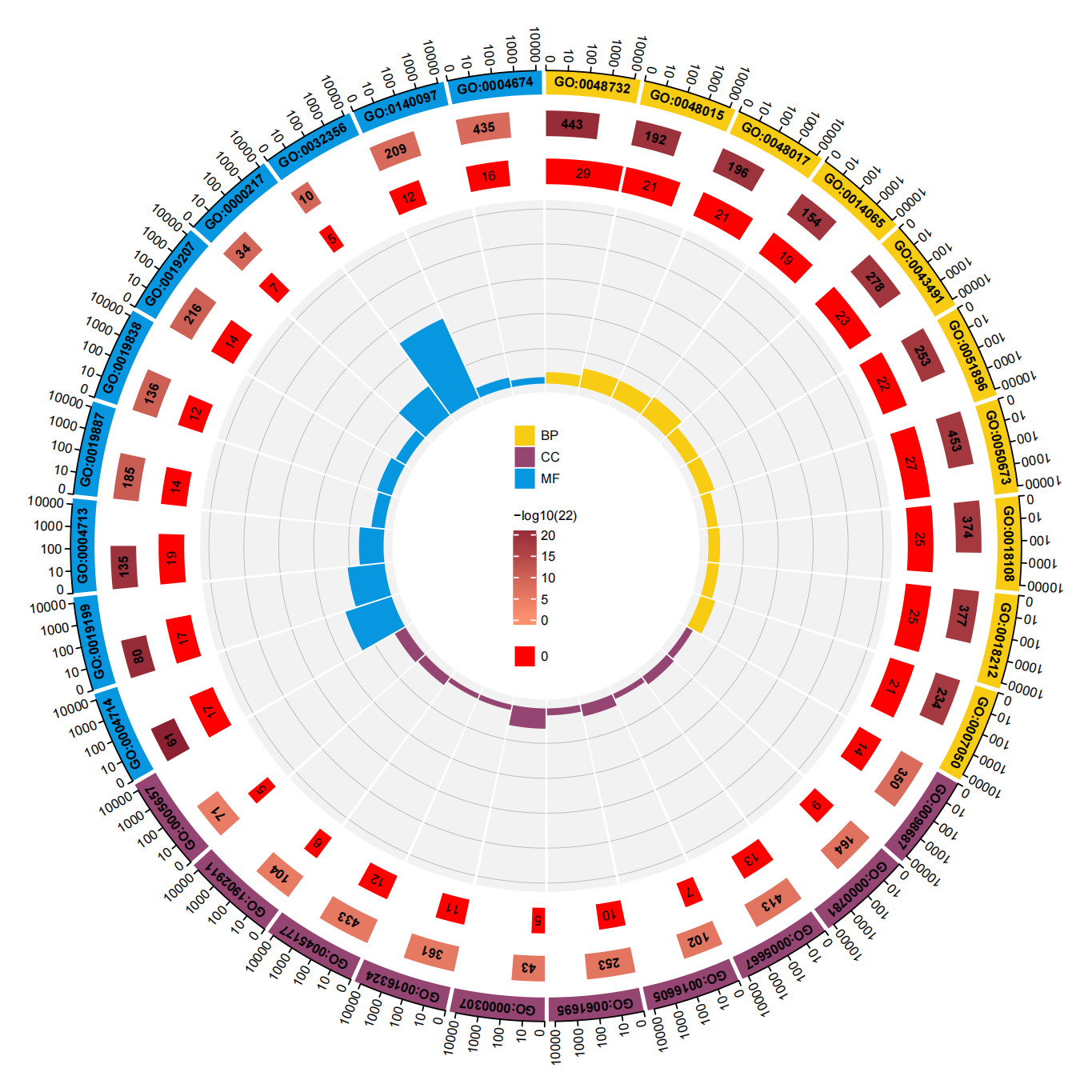 |
ERBB2 non-Ex20突变患者GO富集TOP10 |
重症数据填报网页 2023-12-03 23:30:53 CST
请重症医学科(神经ICU、重症医学科、感染ICU、急诊ICU、外科ICU、CCU)5月28日下午前填好此表发给胡林辉OA。 古科@胡林辉 胡博辛苦您收集填报。同时建立常态化机制,按此表(可以根据实际情况优化)每月、每季度完成时间收集,同时发一份给胡林辉及柏春丽。 每年更换科室负责填报的人。
- 进入3d演示界面
cd e:/py38
.\myenv\Scripts\activate
cd 3d
python app.py- 进入重症数据收集界面
cd e:/py38
.\myenv\Scripts\activate
cd icudatafill
python app.py3d展示平台:http://localhost:8058/
网页展示页面,在Windows本地展示,方便将来移植。 http://localhost:8059/
代码编辑文件夹e:/py38/icudata
mermaid官网:https://mermaid.js.org/syntax/flowchart.html, 在线编辑器https://mermaid.live/ render后按
Ctrl + R刷新才能正常显示。
graph TD
subgraph "Check pathname validity"
B{Check pathname validity}
end
B -->|Invalid Pathname| C(Render 404 page)
B -->|Valid Pathname| D{Check authentication status}
subgraph "If authenticated:"
D -->|Authenticated| E{Redirect or Render Index page}
end
subgraph "If not authenticated:"
D -->|Not Authenticated| F{Render Login or Redirect to Login}
end
E -->|Pathname is /login| G{Redirect to Main Page}
E -->|Pathname is not /login| H(Render Index Page)
F -->|Pathname is /login| I(Render Login Page)
F -->|Pathname is /relogin| J(Render Login Page with relogin)
F -->|Other Pathnames| K(Redirect to Login Page)
- dcc.Location的原理是什么?
在 Dash 中,
dcc.Location组件的原理是监听浏览器 URL 地址的变化,并将这些变化反映到 Dash 应用程序中。具体来说,dcc.Location组件通过 JavaScript 监听浏览器 URL 地址的变化,然后将变化的信息发送到 Dash 应用程序的后端,以便后端能够根据 URL 地址的变化执行相应的逻辑处理。当浏览器的 URL 地址发生变化时,
dcc.Location组件会触发一个名为pathname的属性发生变化的事件。这个事件会被发送到 Dash 应用程序的后端,进而触发与dcc.Location组件相关联的回调函数。在回调函数中,开发者可以根据 URL 地址的变化情况执行相应的逻辑处理,例如根据不同的 URL 地址加载不同的页面内容、执行重定向操作或更新应用程序的状态。
总之,
dcc.Location组件的原理是通过监听浏览器 URL 地址的变化,将这些变化反映到 Dash应用程序中,并触发与其关联的回调函数来执行相应的逻辑处理,从而实现页面内容的动态刷新和路由控制的功能。 在 Dash 中,dcc.Location 组件的 pathname 属性可以用来获取当前 URL 中的路径部分,并且也可以通过修改 pathname 属性来改变 URL 的路径部分。当你在 Dash 应用中修改了 dcc.Location 组件的 pathname 属性时,页面的 URL 地址会相应地发生改变。
这个特性可以用来实现在 Dash 应用中进行页面导航或执行重定向的功能。通过修改 dcc.Location 组件的 pathname 属性,你可以动态地改变页面的 URL 地址,从而实现页面内容的动态刷新和路由控制的功能。
总之,dcc.Location 组件的 pathname 属性可以用来改变 URL 的路径部分,从而实现在 Dash 应用中进行页面导航或执行重定向的功能。
为方便按关键词定位到代码所有py文件,可用以下代码将所有py代码的内容汇总至一个单独的文本文档
import os
def print_py_files_content_with_path(directory):
for root, dirs, files in os.walk(directory):
for file in files:
if file.endswith(".py") and 'checkpoint' not in file:
file_path = os.path.join(root, file)
print(f"File: {file_path}")
with open(file_path, 'r', encoding='utf-8') as f:
lines = f.readlines()
for line in lines:
print(f"{file_path}: {line.rstrip()}") # 在每行内容前加上文件路径
print("\n")
def write_py_files_content_to_file(directory, output_file):
with open(output_file, 'w', encoding='utf-8') as out_f:
for root, dirs, files in os.walk(directory):
for file in files:
if file.endswith(".py") and 'checkpoint' not in file:
file_path = os.path.join(root, file)
out_f.write(f"File: {file_path}\n")
with open(file_path, 'r', encoding='utf-8') as in_f:
lines = in_f.readlines()
for line in lines:
out_f.write(f"{file_path}: {line}") # 将文件内容及文件路径写入到文本文档中
out_f.write("\n")
更改用户
- 导入用户名:views → login.py → 关键行
from config import Departments; for dept in Departments.deptlst - 调整Departments变量:config.py → 关键行
class Departments: - 增加用户感染科ICU:登录管理员账号,增加即可。原有旧账号不用删除,在登录界面看不到。
- 导入用户名:views → login.py → 关键行
更改界面名称:
- 浏览器名称:server.py → 关键行
app.title = '茂名市人民医院重症医学质控数据填报系统' - 登录界面:views → login.py → 关键行
title='全院抗生素查询系统' - 登录后界面:
\views\index.py→fac.AntdText('全院抗生素查询系统',
- 浏览器名称:server.py → 关键行
ICON修改:
icudatafill\server.py: app.favicon = 'favicon.ico'favicon.ico文件在assets文件夹。 PNG转ICO在线:https://favicon.io/favicon-converter/
点击登录按钮后的流:
- 控件位置:
views\login.py: id='login-submit', - 回调代码-Input:
callbacks\login_c.py: Input('login-submit', 'nClicks'), - 回调代码-Output:
callbacks\login_c.py: Output('login-redirect-container', 'children')], - 回调代码-Output实体位置:
views\login.py: html.Div(id='login-redirect-container'), #重定向容器
- 控件位置:
更改首页标签内容:
- 渲染首页:
app.py: views.index.render_content( - 首页tab容器的位置:
views\index.py: id='index-main-content-container', - 首页tab回调的函数:
callbacks\index_c.py: Input('tabs', 'latestDeletePane'), - 改写首页标签内容的位置:
callbacks\index_c.py: 'children': [ # 这里放首页tab的内容
- 渲染首页:
左、右屏对照效率更高。前面开始做了,后面就有模板可用了,所谓万事开头难。
import requests
from bs4 import BeautifulSoup
import re
import pandas as pd
def extract_titles_from_url(url):
response = requests.get(url)
response.encoding = 'utf-8' # 设置编码方式为utf-8
content = response.text
soup = BeautifulSoup(content, 'html.parser')
# 找到所有包含title属性的标签
title_elements = soup.find_all(attrs={"title": True})
# 提取所有title属性的值
titles = [element.get("title") for element in title_elements]
return titles
# 用法示例
url = 'https://www.mmphcrc.com/linhui/assets/upload/0aaacb09-140d-43fb-810f-b488a520fc8c/%E9%87%8D%E7%97%87%E6%95%B0%E6%8D%AE%E5%A1%AB%E6%8A%A5.html'
titles = extract_titles_from_url(url)
codeDict = {}
# 打印所有title属性的值
pattern = r'(.*?)\((P\d+)\)'
for title in titles:
codeMean, code = re.findall(pattern, title)[0]
codeDict[code] = codeMean
small_dict = pd.DataFrame(codeDict.items(), columns=['代码', '内涵'])
small_dict.代码 = small_dict.代码.str.lower()
small_dict.head()
big_dict = pd.read_excel('2023年ncisdc系统【1综合-三级综合医院】填报指标-20230519.xlsx')
big_dict.head()
code_data_structure = (
pd.merge(small_dict, big_dict, left_on="代码", right_on = "字段名")
[['代码', '内涵', '类型', '长度']]
)
code_data_structure.代码 = code_data_structure.代码.str.upper()
code_data_structure.to_csv('代码内涵字典表.csv', index=False)
code_data_structureOutput和Input均可以通过解包的形式写,相同格式的可以这样写,减少代码量。
callbacks\index_c.py: linkDict=[
工作流(溯源):
views\index.py: 'children': Menu.submenuItemsHosp if role == '管理员' else Menu.submenuItemsDeptviews\index.py: from config import Menuconfig.py: class Menu:config.py: submenuItemsDept = [config.py: 'title': '抗生素DDD查询',callbacks\index_c.py: from config import Table, TabItemscallbacks\index_c.py: *TabItems.tabItemChildren.get(title, []) #根据情况追加标签内元素左侧导航栏代码位置:
学科建设评估平台
春节前完成,2024年2月10日
计划运行网页 https://www.mmphcrc.com/linhui/posts/学科建设能力评估系统/学科建设能力评估系统.html
思平:多发时间辅助作业,系统性培养思维
思扬:多发时间早教
- 可以玩,但当我要求做作业的时候,要赶快去做,专心致志地做作业。
- 2023-10-06 12:10 手背打了两次脸,拍了屁股,踢了一脚屁股。从此,爱有缺陷,也许童年有阴影,但仍然要长抓下去。
| 成长型思维 | 固定型思维 |
|---|---|
| 通过努力,智力和能力是可以提升 | 智力和能力是与生俱来的,后天努力不可能带来任何改变 |
| 挑战是学习的好机会 | 遇到挑战说明能力不够 |
| 失败是成功的准备 | 失败说明我天生不是这块料 |
| 从周围人的成功上学习 | 周围人的成功说明我不够好 |
- 严格要求,不要放纵。
- 处理好与同学的关系:Deng的无礼骚扰,语言还是行为,都要针锋相对予以还击。
第4周
电脑浏览器访问培训平台网址https://jnyjs.yuketang.cn,微信扫码登录。
【金山文档】 呼吸机病理生理 https://kdocs.cn/l/ccRGzVUg71FA
文献翻译http://www.mmphcrc.com:8050/pubmed
【金山文档】 Point-by-point-response-to-reviewers-R3 https://kdocs.cn/l/ccObiGqSzq1k
【金山文档】 manuscript-I3A-R3 https://kdocs.cn/l/cfzY9laGYUCf 【金山文档】 Ethics approval https://kdocs.cn/l/cn7ld0yXGLj4
无鼠标纯用键盘连接蓝牙 1. 按Windows图标,通过Tab和上下左右箭头以及回车键进入”设置”。 2. 进入蓝牙设备,Tab键选中三个
.,即...,按回车删除设置,将已连接的3个设备选删除。 3. Tab键选中添加设备,进行添加设备。
Dear Dr Chen,
Re: “Plasma indole-3-aldehyde as a novel biomarker of acute kidney injury after cardiac surgery: a reanalysis using prospective metabolomic data” We are pleased to let you know that your manuscript has now passed through the review stage and is ready for revision. Many manuscripts require a round of revisions, so this is a normal but important stage of the editorial process. Editor comments 1. The ethics approval with reference number PJ2020MI-021-01 is used in another published article, https://www.frontiersin.org/articles/10.3389/fmicb.2023.1119959/full. Please explain in details what happened, and also provide us a copy of the ethics approval document for check. Why patients recruitment period is different in the two articles? 2. That published article should be cited properly in your manuscript, and please describe the difference between this study and published work, and what this study adds to the published work in detail. 3. Why “the exclusion criteria of the two studies differ” considering these are using the same approval document? To ensure the Editor and Reviewers will be able to recommend that your revised manuscript is accepted, please pay careful attention to each of the comments that have been pasted underneath this email. This way we can avoid future rounds of clarifications and revisions, moving swiftly to a decision.
Once you have addressed each comment and completed each step listed below, the revised submission and final file can be uploaded via the link below.
If you completed the initial submission, please log in using the same email address. If you did not complete the initial submission, please discuss with the submitting author, who will be able to access the link and resubmit.
https://submission.springernature.com/submit-revision/26d9784d-19b5-4195-8e85-2a4e3464e304
You can visit https://researcher.nature.com/your-submissions to track progress of this or any other submissions you might have.
CHECKLIST FOR SUBMITTING YOUR REVISION
- Please upload a point-by-point response to the comments, including a description of any additional experiments that were carried out and a detailed rebuttal of any criticisms or requested revisions that you disagreed with. This must be uploaded as a ‘Point-by-point response to reviewers’ file.
Please note that we operate a transparent peer review process, where we publish reviewers’ reports with the article, together with any responses that you make to reviewers or the handling Editor.
Please highlight all the amends on your manuscript or indicate them by using tracked changes.
Check the format for revised manuscripts in our submission guidelines, making sure you pay particular attention to the figure resolution requirements:
https://bmcanesthesiol.biomedcentral.com/submission-guidelines
Finally, if you have been asked to improve the language or presentation of your manuscript and would like the assistance of paid editing services, we can recommend our affiliates, Nature Research Editing Service: https://authorservices.springernature.com/language-editing/ and American Journal Experts: https://www.aje.com/go/springernature
Please note that use of an editing service is neither a requirement nor a guarantee of publication. Free assistance is available from our resources page: https://www.springernature.com/gp/researchers/campaigns/english-language-forauthors
To support the continuity of the peer review process, we recommend returning your manuscript to us within 14 days. If you think you will need additional time, please let us know and we will aim to respond within 48 hours.
Kind regards,
Guangde Tu Editorial Board Member BMC Anesthesiology
Reviewer Comments:
Reviewer 3 I have no further comments.
Reviewer 1 As a reviewer, I have carefully considered the contents of this article. The authors appear to have engaged in adequate discussion and revisions in response to the reviewers’ feedback. However, there are some minor points of critique as follows.
As the authors have indicated in the title, they have re-analyzed some of the data from a previously published paper. Therefore, a reference to the published paper should be included. (No. PJ2020MI-021-01). “Li et al, Evaluation of the contribution of gut microbiome dysbiosis to cardiac surgery-associated acute kidney injury by comparative metagenome analysis, Front. Microbiol., 17 March 2023, Sec. Systems Microbiology (The study was performed according to the guidelines in the Helsinki Declaration, and the local ethics committee approved the study with no. PJ2020MI-021-01 and no. KY-Q-2021-109-04.)”
In addition, the exclusion criteria of the two studies differ. Is there a reason for this difference? This paper: “The exclusion criteria were pre-existing advanced CKD (end-stage renal disease, renal transplantation), age of >80 years, a pre-existing malignant tumour and refusal to consent. To assess the ability to predict AKI, patients who had been exposed to AKI from a prior operation were also excluded.” Front. Microbiol., 17 March 2023: “Reasons for exclusion were as follows: (1) age under 18 years; (2) received systemic antibiotic treatment within 48 h before admission; (3) a history of nephrectomy or ESRD or renal transplantation; (4) died within 24 h after cardiovascular surgery; (5) lack of preoperative fecal samples. Written informed consent was obtained from the patients before the surgery.”
老师署名单位为:Department of Critical Care Medicine, Shenzhen People’s Hospital (The Second Clinical Medical College,Jinan University; The First Affiliated Hospital, Southern University of Science and Technology), Shenzhen 518020, Guangdong, China.
深圳市人民医院关于学术论文单位标注规范的通知
发布科室:科研科 发布时间:2021-07-16
已阅人数:419
各科室、龙华分院、坂田院区:
为确保深圳市人民医院科研信息统计、科研成果认定和科研业绩奖励的准确性,本院所有科研人员及课题组成员,包括访问学者及访问学生,发表论文的单位应依照此说明标注。
因标注不规范,导致成果无法通过各种科学文献检索数据库搜索而被收录进深圳市人民医院科研管理系统的,其成果在各类政府统计和科室、个人考核中,医院原则上不予计入。如情况特殊,需提出申请,由医院学术委员会批准,方可计入。
一、所有论文署名要求统一标注为:
中文:
深圳市人民医院(暨南大学第二临床医学院,南方科技大学第一附属医院)眼科 广东深圳 518020
英文:
Department of xxx, Shenzhen People’s Hospital (The Second Clinical Medical College,Jinan University;The First Affiliated Hospital, Southern University of Science and Technology), Shenzhen 518020, Guangdong,China
二、院系、科研平台等建议分级标注为:
依托我院建设的国家级、省部级、市级及其他科研创新平台(重点实验室、工程中心、工程技术中心)、本院各科研机构,需首选按照相关部门规定的成果验收考核要求署名。 该类科研创新平台原则上不允许以一级单位独立标注,鼓励并列标注。
中文范例1:
深圳市干细胞研究与临床转化重点实验室
英文范例1:
Shenzhen Key Laboratory of Stem Cell Research and Clinical Transformation,Shenzhen People’s Hospital (The Second Clinical Medical College,Jinan University;The First Affiliated Hospital, Southern University of Science and Technology), Shenzhen 518020, Guangdong,China
三、 并列标注
因相关部门考核要求,鼓励并列标注,顺序可根据相关部门考核要求确定。
中文范例1:
单位1.深圳市人民医院深圳市呼吸疾病研究所 广东深圳 518020
单位2. 深圳市人民医院深圳市呼吸疾病重点实验室 广东深圳 518020
英文范例1:
1. Shenzhen People’s Hospital,Shenzhen Institute of Respiratory Diseases, Shenzhen 518055, Guangdong,China
2. Shenzhen key Laboratory of Respiratory Diseases, Shenzhen People’s Hospital (The Second Clinical Medical College,Jinan University;The First Affiliated Hospital, Southern University of Science and Technology),Shenzhen 518055, Guangdong,China
四、 作者署名及ORCID
作者英文署名应按照期刊要求的格式进行署名,若期刊无规定的,则应使用作者姓名中英文全拼。 通讯作者(corresponding author)应根据不同期刊的要求,在作者姓名后标注,同时提供ORCID,以便检索。
五、本说明由科研科负责解释。
深圳市人民医院科研科
2021年7月16日
结构化病历
- 【金山文档】 病历书写与管理基本规范(2022版) https://kdocs.cn/l/ctphKT1GHjqD
- 【金山文档】 病历书写与管理基本规范(2022版)(OCR) https://kdocs.cn/l/cgqsrCcpUziG
- 【金山文档】 病历书写与管理基本规范(2022版)(OCR-Bookmark) https://kdocs.cn/l/chYbOvCqCQ1I
【金山文档】 首次病程记录-1-大颗粒度版本 https://kdocs.cn/l/crVwbFlgpOih 【金山文档】 首次病程记录-1-大颗粒度版本 https://kdocs.cn/l/cpi5IjE2wsJc
https://github.com/hulihuihong/MyRibbonAddIn
文件放在
桌面/知情同意书文件夹
| 重大疾病代码 | 名称 |
|---|---|
| 2-139-1 | 无 |
| 2-77-1 | 肝硬化 |
| 2-77-2 | 恶性肿瘤 |
| 2-77-3 | 慢性肾功能衰竭(尿毒症期) |
| 2-77-4 | 再生障碍性贫血 |
| 2-77-5 | 系统性红斑狼疮 |
| 2-77-6 | 规定项目组织器官(肝脏、肾脏、心脏)移植 |
| 2-77-7 | 心脏病合并心功能不全Ⅲ级及以上 |
| 2-77-8 | 地中海贫血 |
| 2-77-9 | 儿童先天性心脏病 |
| 2-77-10 | 白血病 |
| 2-77-11 | 原发性血小板增多症 |
| 2-77-12 | 多器官功能衰竭 |
| 2-77-13 | 重型颅脑损伤 |
| 2-77-14 | 特重度烧伤 |
| 2-77-15 | 骸关节置换术 |
| 2-77-16 | 全身多处骨折 |
| 2-77-17 | 骨髓增生异常综合症 |
| 诊疗方式代码 | 名称 |
|---|---|
| 2-70-0 | 保守治疗 |
| 2-70-1 | 传统手木 |
| 2-70-2 | 微创手术 |
| 2-70-3 | 介入治疗 |
| 2-70-4 | 透析及血液过滤 |
| 2-70-5 | 透析治疗 |
| 2-70-6 | 干细胞移植 |
| 2-70-7 | 机械通气 |
| 2-70-8 | 超声乳化 |
| 2-70-9 | 玻璃体切割 |
| 2-70-10 | 适形放疗 |
| 2-70-11 | 适形调强放疗 |
| 2-70-12 | 腹水回输术 |
| 2-70-13 | 修复手术 |
| 2-70-14 | 体外碎石 |
| 2-70-15 | 容积调强 |
| 2-70-16 | 血液净化 |
| 2-70-17 | 机械持续降温 |
| 2-70-18 | 器官移植 |
| 2-70-19 | 冠脉造影 |
| 2-70-20 | 同期放化疗 |
| 2-70-21 | 生物免疫治疗 |
| 2-70-22 | 双侧手术 |
| 2-70-23 | 化疗药物治疗 |
| 2-70-24 | 保守无痛治疗 |
| 2-70-25 | 化疗联合靶向治疗 |
| 2-70-26 | 高强度聚焦超声治疗 |
| 2-70-27 | 闭合性复位 |
| 2-70-28 | 溶栓治疗 |
| 2-70-29 | 剖宫产 |
| 2-70-30 | 人工耳蜗 |
| 2-70-31 | 机器人辅助手术 |
| 2-70-32 | 人工肝治疗 |
| 2-70-33 | 放射治疗 |
| 2-70-34 | 肺泡灌洗 |
| 2-70-35 | 双眼手术 |
| 2-70-36 | 高压氧治疗 |
| 2-70-37 | 眼科联合手术 |
| 2-70-38 | 特别药物 |
| 2-70-39 | 手术伴热灌注治疗 |
| 2-70-40 | 联合手术 |
| 2-70-41 | 容积调强联合靶向治疗 |
| 2-70-42 | 阴道分娩 |
试验代码已入Python学习笔记。
JTIM引用
Oral Microbiota and Porphyromonas Gingivalis Kgp Genotypes Altered in Parkinson’s Disease with Mild Cognitive Impairment
群公告 成果登记流程:登录广东省科技业务管理阳光政务平台https://pro.gdstc.gd.gov.cn/egrantweb/(没有账号的请联系科教科注册),点击”过程管理”–“成果登记申请或查询”,点击”新增项目成果登记”并按提示填写各项数据,包括上传结题验收材料等。填写完毕提交审核并将纸质版材料一式一份交科教科,通过科技局审核及公示后(约2个月),可拿到成果登记证书。 备注:不管是省的还是市的项目,都是登录省平台进行成果登记
密码是12345或1234
联系人应用 → 分享 → 全选 →微信分享
软著申报
网址:https://register.ccopyright.com.cn/real.html#/realApplyPeople 账号:18666818197 密码:hjzzq@2922581
2023年11月
第1周
要批量生成文档,可以使用Word的邮件合并功能,具体步骤如下:
准备好邮件模板:在Word中打开一个空白文档,输入需要包含的固定文本,以及需要插入邮件合并数据的位置,例如收件人姓名、地址、电话等。
添加邮件合并数据源:从Word菜单栏中选择”邮件ings”选项卡,在”开始邮件合并”组中选择”选择收件人”,可以从已有的Excel或Access文件中导入数据,也可以手动输入数据源。注意:数据源中的字段名称必须与邮件模板中的标记对应。
添加邮件合并字段:在邮件模板中插入邮件合并字段,例如”<<收件人姓名>>“,这些字段会在邮件合并时自动替换为数据源中的实际数据。
预览邮件合并结果:点击”预览结果”按钮,可以查看每个收件人的具体信息在邮件模板中的表现形式。
完成邮件合并:当确认邮件合并结果无误后,选择”完成合并”按钮,可以将合并结果输出到一个新的Word文档中,每个收件人对应一个独立的文档。
批量保存文档:点击”文件”菜单,选择”另存为”,在弹出的对话框中选择保存位置和文件名,保存文档即可。
通过以上步骤,可以利用Word的邮件合并功能批量生成文档。
扣款日:2023年11月1日,60日内免息。
您尾号5294的龙卡信用卡11月01日22:09消费/预付款2790.00美元,实际消费金额以入账结算币种金额为准。因额度不足,本次交易已为您提供超额度用卡服务,超额470.26元,如需继续用卡,请您尽快还款。[建设银行] 原文:【金山文档】 I3A-AKI https://kdocs.cn/l/chFGYBmlUxRE 还差出版社发回的发票、pubmed上的检索。 发票:【金山文档】 invoice for i3a https://kdocs.cn/l/crpq5cuDkRlD @ 以后留意发票名字要写茂名市人民医院医生的名字,不要写通讯的名字。
根据绩效考核工作要求,今年1-9月临床科室国考指标数据已OA发给各科科主任和绩效员,请针对1-9月不达标的指标写份整改报告,于11月8日(下周三)前完成整改报告,经科室主任审阅后发回绩效办李伟浩OA,谢谢。
重症医学科三区—2023年1-9月临床科室绩效考核指标完成情况
【通知】: 为了更快更好的适应医保DIP支付方式改革,根据医院工作要求,请各临床科室在本周五(11月3日)下午下班前,将本科室2023年1-10月收治的数量排名前10病种,逐一对标《茂名市DIP病种目录库》中的DIP分值,填报《各科室收治前10病种统计表格》报质控科张沛荣OA。拟定于11月8日-10日开展联合现场抽查。【质控科、医保科、医务科2023.11.2】
【金山文档】 附1:茂名市DIP病种目录库(核心综合基层)https://kdocs.cn/l/cjQ5WgvYAUuL 【金山文档】 重症医学科三区2023年1-10月收治的数量排名前10病种分值统计 https://kdocs.cn/l/csVigbq6gSVe
美食收集
黑名单
- 阳光玫瑰葡萄不是越大越贵越好吃:28元一斤买了2斤,又酸又烂。
白名单
- 路达老广记:猪杂粥,肠粉,量大,好吃。
第2周
每个病人整理出一个PDF,挺好处理。导出文本,正则是关键。
matplotlib学习
jupyter旧资源断舍离
第3周
Letter to editor
双优
Python教育
照片和视频入库
思平学习
GCP
科研培训
用quarto做
简历制作
- 简历模板制作
孟德尔随机论文
PPT制作
12月16日(周六) 分会场九【二层会议室5】 16:30 – 16:50 分会场会议 肾脏替代治疗 (Renal Replacement Therapy) 决策流程 (Decision-making algorithm) 讲者
连续性肾替代治疗规范化治疗流程 https://mp.weixin.qq.com/s/3cOabJp2d70nYP3jtrAdkg
陈纯波教授讲决策,可以参考(PMID: 28119408; 34911731)
AKI背景
- AKI发病病机制
CRRT对AKI预后的影响
启动的时机
结束的时机
总结
